Resources
- Breast cancer GWAS SNPs
- A list of breast cancer GWAS SNPs, their high LD SNPs and coordinates can be obtained from here.
- The complete GWAS annotation catalog (April 2018 version) used in our database can be downloaded from here. The latest version is available on the EBI website.
- WashU Epigenome Browser
- We use the WashU Epigenome Browser interface for showing data of different types.
- The source code for building the interface using Javascript and CSS can be found here.
- TCGA data
- Gene expression quantification (HTSeq - FPKM, open access) data was downloaded from the GDC data portal.
- Raw data such as RNA-seq aligned reads and patient cohort genotypes was also obtained from the GDC data portal after receiving permission to access controlled data.
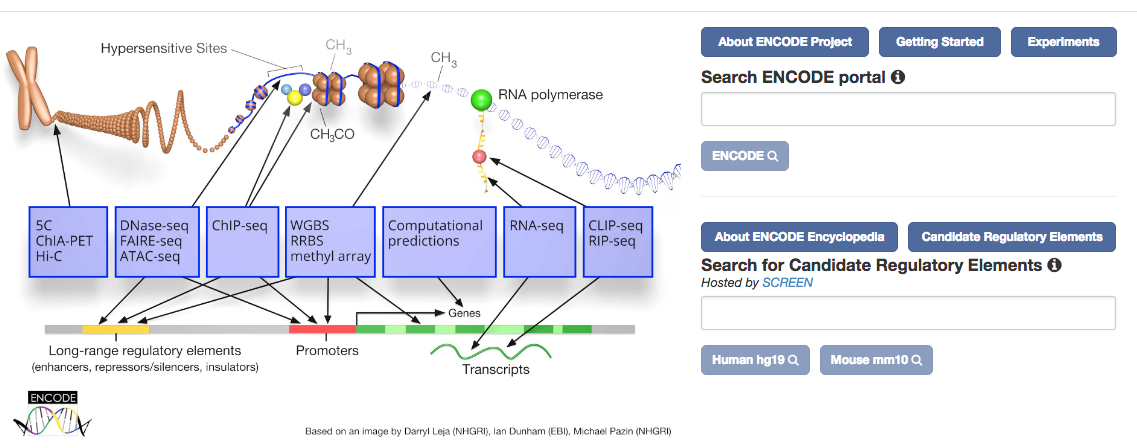
- ENCODE data
- ChIP-seq narrowPeak and broadPeak files were downloaded (April 2018 update) from the ENCODE portal.
- The bam files are also downloaded from the ENCODE portal and sliced before using in the genome browser when relevant.
- Motif databases
- The following motif databases are included in the motif analysis: JASPAR, TRANSFAC, HOCOMOCO, HOMER, hPDI, FactorBook, Jolma.
- Motif logos are generated using the R package seqLogo.